This post explains why it is crucial to annotate your pathology samples consistently and provides you with downloadable annotation guidelines.
The sources of variability
Digital tissue diagnostics utilizes the power of image analysis to extract quantitative information from conventional pathology slides. However, it is a complex process consisting of many steps that can potentially lead to erroneous results. Before the sample is subjected to digital analysis the tissue must be collected and preserved, embedded in paraffin, sectioned onto glass slides, subjected to antigen retrieval, stained using chemical, immunohistochemical (IHC) or immunofluorescent (IF) techniques and then digitized with a slide scanner. Each of the steps has inherent preanalytical or analytical variability, which on top of tumor heterogeneity and biological variability, makes it challenging to generate consistent input for image analysis.
Additionally, tissue diagnostics suffers from intra-observer variability when it comes to the interpretation of the sample. Some of the challenges here are diagnostic drift – a gradual change of nomenclature and severity grading of lesions within a single study, and chronological bias which is the evolution of a grading system with increasing experience (detection of more subtle changes with time).
Various publications cover the problem of inter-observer variability and talk about how much pathologists differ between each other when it comes to IHC interpretation. This is an area where digital analysis can help solve the problem. It makes the IHC interpretation as quantitative and as objective as possible. Nevertheless, pathologist input is necessary to accurately identify the region of interest, such as the area of the tumor, which usually initiates the digital image analysis workflow.
Digital image analysis workflow
The digital image analysis workflow consists of continuous communication and maintenance of a feedback loop between the pathologist who interprets tissue in its biological context, and the computer scientist, who translates the structures and colors into quantifiable objects of different classes. The first step of this communication is the manual delineation of the tumor mass on the digitized tissue slide by a pathologist, also known as region annotation. As this input will later be used to generate numerical results on which clinical decisions may be made, there is little room for inter- or intra-observer variability.
Trained as morphologists, pathologists are very competent in recognizing and classifying the tissue components, such as different types of tumors, inflammation, necrosis, fibrosis, etc. This knowledge is indispensable to assure meaningful, quantitative image analysis. However, when faced with a quantitative task such as IHC interpretation, they are often challenged, as the human eye does not have the capacity to accurately quantify the amount of stain present in the sample. The most comprehensive approach to tissue diagnostics is to use a pathologist’s expertise in recognizing and annotating the relevant regions and the power of a digital solution to quantify the IHC within these regions.
To minimize the discrepancies between pathologists in region annotations, to address the technical needs of digital image analysis and to make the downstream analysis reproducible, we created region annotation guidelines. The use of an annotation software with the possibility of annotation class preselection together with the guidelines minimizes the diagnostic drift and inconsistencies in the nomenclature of the regions of interest. The annotation guidelines are what helps you stay consistent throughout your project and across different projects.
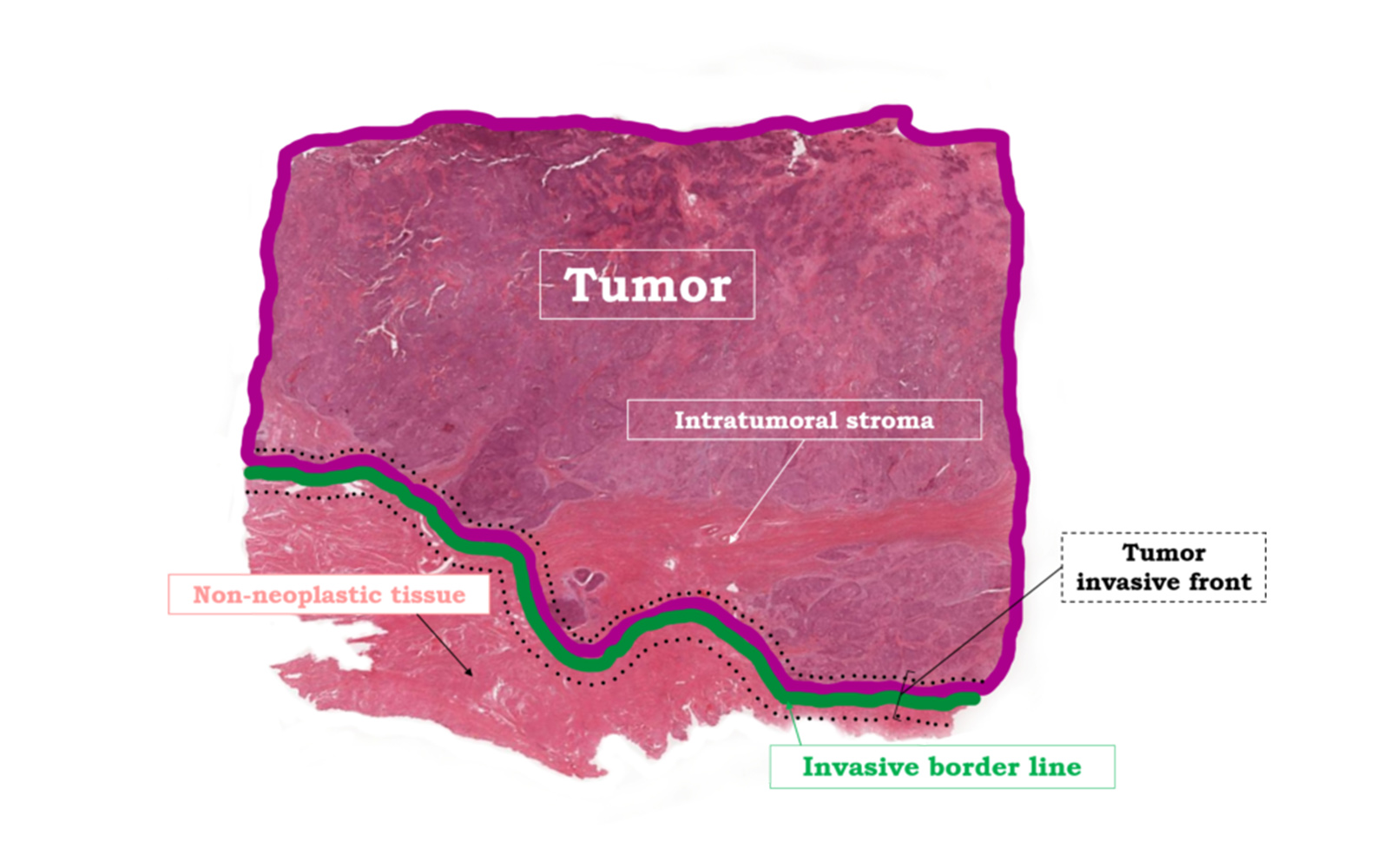
The region of interest: Tumor, Invasive Border Line and Invasive Front
The most relevant regions, which require a clear understanding and definition, are TUMOR, INVASIVE BORDER LINE, and INVASIVE FRONT. These terms, as intuitive as they are to every trained pathologist, need to be defined in an exact way to ensure proper communication with all other experts involved in the process of image analysis.
For this purpose,
TUMOR
is defined as the main mass of neoplastic cells together with the supporting intratumoral stroma present on the slide.
INVASIVE BORDER LINE
is the border line between the outer neoplastic cells and the surrounding non-neoplastic tissue and
INVASIVE FRONT
is the area of a certain width on both sides of the invasive border line, also known as the tumor-host interface or invasive margin. In this area, the tumor invades the surrounding non-neoplastic tissue and the host immune cells are invading the tumor from the outside. In addition to the tumor itself, the invasive front is an area of high immuno-oncological interest. This is where many relevant immune cell interactions take place. INVASIVE FRONT in vivo is a three-dimensional area. Here we apply this concept to a two-dimensional slide to obtain maximal annotation consistency.
The annotation guidelines include:
- Definitions of relevant regions
- Graphical representation of all relevant regions
- Indications on how to annotate the relevant regions
- Distinction of which regions should be annotated manually, and which should be generated automatically by a software
If you want to keep consistency within and across your digital pathology image analysis projects, you can subscribe and download the annotation guidelines here:
This is a great resource to share with both the pathologists and computer scientists on your team. Use them as they are or adapt to your own project. Use annotation guidelines consistently and you will not have to worry if the results of your projects are distorted by the way you annotate your samples.












Comments are closed.